
Contact
- (210) 567-3976
- kadosh@uthscsa.edu
Department
Microbiology, Immunology & Molecular GeneticsKadosh, David, Ph.D.
Professor
Personal Statement:
As a graduate student at Harvard University, Dr. Kadosh’s studies focused on transcriptional regulation in the model yeast Saccharomyces cerevisiae. He was one of the first to demonstrate that recruitment of histone deacetylases to target promoters causes transcriptional repression; this fundamental mechanism is conserved from yeast to mammals and is known to play a role in a variety of disease processes.
As a postdoctoral fellow at the University of California, San Francisco, he shifted his focus to the major human fungal pathogen Candida albicans where he identified and characterized several key regulators of C. albicans filamentous growth and virulence. At the University of Texas Health Science Center San Antonio, the Kadosh laboratory has made a number of important contributions towards the understanding of how specific transcriptional and translational mechanisms control morphology, virulence, biofilm formation as well as the response to host stresses in C. albicans as well as other pathogenic Candida species.
They have shown that all three C. albicans morphologies are determined by a common dosage-dependent transcriptional mechanism and, using genomic and bioinformatic analysis, identified transcriptional profiles and global gene expression patterns specifically associated with C. albicans pseudohyphal and hyphal morphologies.
His laboratory also provided the first demonstration that a major C. albicans virulence property, morphology, is controlled by a 5’ UTR-mediated translational efficiency mechanism as well as evidence that certain key C. albicans morphological and biofilm regulatory functions show partial evolutionary conservation in non-albicans Candida species. Recently, the Kadosh laboratory has used ribosome profiling to define the first genome-wide translational profiles of C. albicans cells that are responding to antifungal treatment and undergoing morphogenesis.
The Kadosh lab has also established several highly productive collaborations with members of the large and internationally recognized San Antonio Center for Medical Mycology (http://www.sacmm.org/).
Education
Ph.D., Harvard University (1998)
Research
Dr. Kadosh’s laboratory studies the major human fungal pathogen Candida albicans Although present as a commensal in the digestive tract of most healthy people, C. albicans is also capable of causing a wide variety of systemic and mucosal infections.
Immunocompromised individuals such as AIDS patients, organ transplant recipients, cancer patients undergoing chemotherapy, and recipients of artificial joints and prosthetic devices are particularly susceptible to infection. C. albicans is also a highly evolved fungal pathogen, capable of invading nearly every organ and tissue in the human body; indeed, there are no known reservoirs for C. albicans outside the mammalian host.
How is C. albicans able to function so effectively as a pathogen? This organism possesses a number of properties which contribute to its virulence, including secretion of degradative enzymes and adhesion to host cells. Research in our laboratory focuses on one such virulence property, the ability to undergo a reversible morphological transition from yeast (single oval cells) to pseudohyphal and hyphal filaments (elongated cells attached end-to-end, see Figure 1). Previous studies have shown that this transition is important for virulence and can occur in response to a wide variety of host inducing signals. We also study how C. albicans can form biofilms and tolerate antifungal treatment as well as a wide variety of stress conditions in the host environment.
We use a variety of genetic, genomic, biochemical and molecular biology approaches to gain a better understanding of the mechanisms that control the C. albicans morphological transition, biofilm formation, and stress responses in response to specific host environmental cues.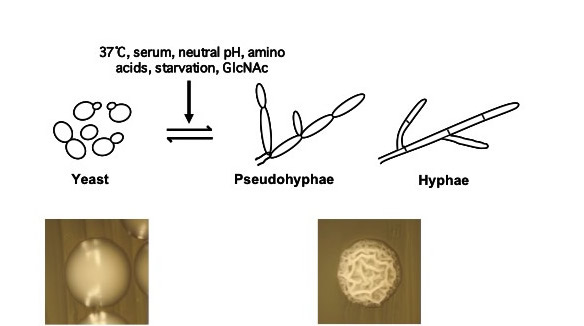

Transcriptional and Translational Mechanisms Important for Determination of Candida albicans Morphology and Virulence
We have identified a filament-specific transcriptional regulator, UME6, which plays an important role in promoting C. albicans hyphal filament extension, virulence and biofilm formation. UME6 is a component of the C. albicans filamentous growth program and appears to promote hyphal extension by maintaining the level and duration of expression of filament-specific transcripts in response to host environmental cues.
Constitutive high-level expression of UME6 is sufficient to drive complete hyphal formation under non-filament-inducing conditions in vitro and causes a significant increase in hyphal formation, tissue invasion and virulence during infection in vivo.
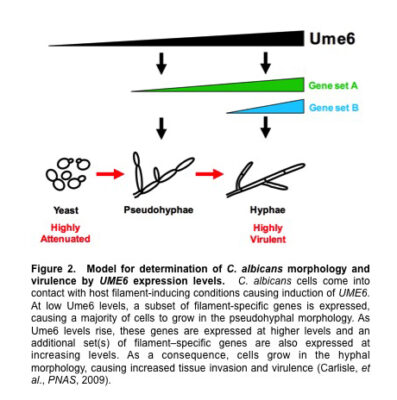
These results provided strong evidence that the C. albicans hyphal morphology plays a specific important role in virulence. Strikingly, we have found that as UME6 levels increase C. albicans shifts morphology from yeast to pseudohyphae to hyphae; there is an accompanying increase in both the number of filament-specific genes expressed as well as their level of induction.
These results indicate that UME6 levels alone are sufficient to determine C. albicans morphology and suggest that all three morphologies are controlled by a common dosage-dependent transcriptional mechanism (Figure 2). Whole-genome transcriptional profiling experiments suggested that genes associated with pseudohyphal growth represent a subset of those associated with hyphal growth and are generally expressed at lower levels.
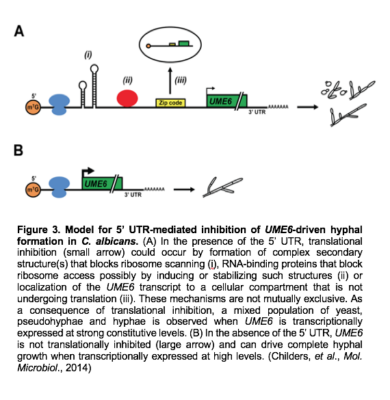
We have also demonstrated that C. albicans UME6 possesses one of the longest 5’ untranslated regions (UTRs) identified in yeast to date. This 5’ UTR functions to inhibit translational efficiency of UME6 and reduces the ability of UME6 expression to drive complete hyphal growth (Figure 3). Interestingly, the level of translational inhibition is modulated by host filament-inducing conditions.
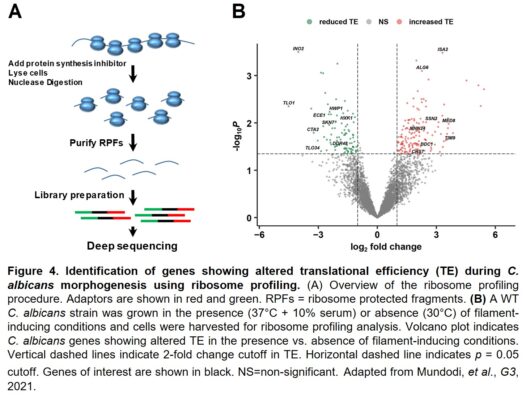
Finally, we have used genome-wide ribosome profiling to determine the complete sets of C. albicans genes that showed altered translational efficiency during morphogenesis as well as in response to fluconazole, a mainline antifungal treatment (Figure 4). Interestingly, we have observed significant differences in genes that are regulated at the translational vs. transcriptional levels. In addition, many virulence factors that are strongly induced at the transcriptional level during morphogenesis show reduced translational efficiency, suggesting that a translational fine-tuning mechanism functions to control filament-specific gene expression.
We are currently using a variety of approaches to further investigate both transcriptional and translational mechanisms important for determination of C. albicans morphology, virulence and virulence-related processes.
Identification and Characterization of Novel C. albicans Virulence Factors
Genes in the C. albicans filamentous growth program, as well as genes that show altered translational efficiency during morphogenesis are involved in a wide variety of biological processes, several of which had not previously been implicated in virulence. Several of these genes are unique to C. albicans, suggesting the possibility that they may be important for novel virulence mechanisms. Efforts are currently underway to further characterize these genes and determine the precise roles that they play in allowing C. albicans to establish and maintain infection in host tissues. Genes involved in translational mechanisms associated with C. albicans virulence properties are of particular interest. While many effective antibiotics are known to target bacterial translation mechanisms, very few antifungals are available to target the translation machinery.
Evolution of Morphology and Virulence in Candida Species
Approximately 50% of all Candida infections are caused by C. albicans and the remainder can be attributed to a variety of less pathogenic non-albicans Candida species (NACS). NACS are believed to be less pathogenic than C. albicans due to a reduction in adherence to host cells, biofilm formation, secretion of degradative enzymes and filamentation as well as increased sensitivity to cell stresses.
However, certain NACS have shown increasing resistance to antifungals and are emerging as important fungal pathogens in the clinic. Considerably little is known about the evolution of regulatory mechanisms important for controlling morphology and biofilm formation in pathogenic Candida species. We have defined specific optimal environmental conditions that induce filamentation in three NACS which are already starting to pave the way for additional work in the field.
We have also demonstrated that an evolutionary weakness in the induction of conserved orthologs of C. albicans filament-specific target genes can be partially overcome by high-level expression of UME6 orthologs. These findings are significant because they suggest that C. albicans morphological and biofilm regulatory functions are partially conserved in NACS as well as a novel hypothesis that filamentation ability may have evolved with strength of filament-specific gene expression.
Awards & Accomplishments
| Year | Name |
|---|---|
| 2015-12 | Faculty of the Year, UT Health San Antonio Department of Microbiology, Immunology & Molecular Genetics |
| 2014-10 | Institute for Integration of Medicine and Science Clinical and Translational Science Award |
| 2014-09 | San Antonio Life Sciences Institute Innovation Challenge Award |
| 2011-04 | Finalist, Burroughs Wellcome Fund Investigators in the Pathogenesis of Infectious Disease Award |
| 2009-07 | Voelcker Young Investigator Award |
| 2009-01 | Proceedings of the National Academy of Sciences USA Recruited Author |
| 2008-03 | Burroughs Wellcome Fund ASM Junior Faculty Travel Award |
| 2006-07 | UTHSCSA Executive Research Committee New Investigator Award |
| 2002-04 | Finalist, Burroughs Wellcome Fund Career Award in the Biomedical Sciences |
| 1998-10 | Damon Runyan Cancer Research Fund Postdoctoral Fellow, UCSF |
| 1995-05 | Albert J. Ryan Fellow, Harvard Medical School |
| 1992-09 | National Science Foundation Predoctoral Fellow, Harvard Medical School |
Affiliations
- American Society for Microbiology
- International Society for Human and Animal Mycology
- Medical Mycology Society of the Americas
- San Antonio Center for Medical Mycology
- Cancer Therapy and Research Center, UTHSCSA
- American Heart Association
Publications
- *Choudhary, S., *Mundodi, V., Smith, A.D., Kadosh D. Genome-wide translational response of Candida albicans to fluconazole treatment. Microbiol. Spectr. 11(5):e0257223. (2023)
- Sharma, C., Kadosh, D. Post-transcriptional control of antifungal resistance in human fungal pathogens. Crit. Rev. Microbiol. 49(4):469-484. (2023)
- Xu, C., Kadosh, D., Sun, D., Zeng, G., Gao, Y.Z. Editorial: Omics-originated exploration of pathogenic patterns and molecular mechanisms in human and animal fungal pathogens. Front. Microbiol. 14:1243709. (2023)
- Sharma, C., Kadosh, D. Perspective on the origin, resistance, and spread of the emerging human fungal pathogen Candida auris. PLoS Pathog. e1011190. (2023)
- Kadosh, D. Rapid proliferation compensates for defective filamentation in Candida albicans pathogenesis. Trends Microbiol. 29(10):867-868. (2021)
- *Mundodi, V., *Choudhary, S., Smith, A.D., Kadosh. D. Global translational landscape of the Candida albicans morphological transition. G3 (Bethesda) 11(2):jkaa043. (2021)
Selected as a featured article - Kadosh, D., Mundodi, V. A Re-evaluation of the relationship between morphology and pathogenicity in Candida species. J. Fungi 6, 13 (2020)
- *Banerjee M, *Lazzell A.L., Romo J.A., Lopez-Ribot J.L., Kadosh D. Filamentation is associated with reduced pathogenicity of multiple non-albicans Candida species. mSphere. Oct 16;4(5) (2019).
Featured as “Editor’s Pick” article - Romo, J.A., Zhang, H., Hong, C., Kadosh, D., Koehler, J., Saville, S.P., Wang, Y., Lopez-Ribot, J.L. Global transcriptomic analyses of the Candida albicans response to treatment with a novel inhibitor of filamentation. mSphere Sep 11;4(5). doi: 10.1128/mSphere.00620-19 (2019).
Featured as “Editor’s Pick” article - Kadosh, D. Regulatory mechanisms controlling morphology and pathogenesis in Candida albicans. Curr. Opin. Microbiol. 52:27-34 (2019).
- Vipulanandan, G., Herrera, M., Wiederhold, N., Li, X., Mintz, J., Wickes, B.L., Kadosh, D. Dynamics of mixed-Candida species biofilms in response to antifungals. J. Dent. Res. 97(1):91-98 (2018).
- Kadosh, D. Control of Candida albicans morphology and pathogenicity by post-transcriptional mechanisms. Cell. Mol. Life Sci. 73(22):4265-4278 (2016).
-
Kadosh, D., Najvar, L.K., Bocanegra, R., Olivo, M., Kirkpatrick, W.R., Wiederhold, N.P., Patterson, T.F. Effect of antifungal treatment in a diet-based murine model of disseminated candidiasis acquired via the gastrointestinal tract. Antimicrob. Agents Chemother. 60(11): 6703-6708 (2016).
-
Albataineh, M.T. and Kadosh, D. Regulatory roles of phosphorylation in model and pathogenic fungi. Med. Mycol. 54(4):333-352 (2016).
-
Childers, D.S. and Kadosh, D. Filament condition-specific response elements control the expression of NRG1 and UME6, key transcriptional regulators of morphology and virulence in Candida albicans. PLoS One 10(3):e0122775 (2015).
-
Vautier, S., Drummond, R.A., Chen, K., Murray, G.I., Kadosh, D., Brown, A.J., Gow, N.A., MacCallum, D.M., Kolls, J.K., Brown, G.D. Candida albicans colonization and dissemination from the murine gastrointestinal tract: the influence of morphology and Th17 immunity. Cell. Microbiol. 17(4):445-450 (2015).
Featured as “Editor’s Choice” article -
Albataineh, M.T., Lazzell, A., Lopez-Ribot, J.L. and Kadosh, D. Ppg1, a PP2A-type protein phosphatase, controls filament extension and virulence in Candida albicans. Eukaryot. Cell 13(12):1538-1547 (2014).
-
Childers, D.S., Mundodi, V., Banerjee, M., Kadosh, D. A 5’ UTR-mediated translational efficiency mechanism inhibits the Candida albicans morphological transition. Mol. Microbiol. 92(3):570-585 (2014). Faculty of 1000 “recommended”
-
Kadosh D. Shaping up for battle: morphological control mechanisms in human fungal pathogens. PLoS Pathog. 9(12):100379-100379 (2013).
-
Kadosh D., Lopez-Ribot, JL. Candida albicans: adapting to succeed. Cell Host Microbe 14(5):483-485 (2013).
-
Lackey E, Vipulanandan G, Childers DS, Kadosh D. Comparative evolution of morphological regulatory functions in Candida species. Eukaryot. Cell 12 (10):1356-1368 (2013).
Featured as a Spotlight article, Eukaryot. Cell 12(10):1315 (2013). -
Carlisle, P.L. and Kadosh, D. A genome-wide transcriptional analysis of morphology determination in Candida albicans. Mol. Biol. Cell 24(3):246-260 (2013).
-
Banerjee, M., Uppuluri, P., Zhao, X.R., Carlisle, P.L., Vipulanandan, G., Villar, C.C., Lopez-Ribot, J.L., Kadosh, D. Expression of UME6, a key regulator of Candida albicans hyphal development, enhances biofilm formation via Hgc1- and Sun41-dependent mechanisms. Eukaryot. Cell 12(2):224-32 (2013).
-
*Thompson, D.S., *Carlisle, P.L. and Kadosh, D. Coevolution of morphology and virulence in Candida species. Eukaryot. Cell 10(9):1173-1182 (2011).
-
Carlisle, P.L. and Kadosh, D. Candida albicans Ume6, a filament-specific transcriptional regulator, directs hyphal growth via a pathway involving Hgc1 cyclin-related protein. Eukaryot. Cell 9:1320-1328 (2010).
Featured as a Spotlight article, Eukaryot. Cell 9:1299 (2010).
Featured in the Journal Highlights section of ASM Microbe magazine, Microbe 5(10): 439 (2010). -
Uppuluri, P., Chaturvedi, A.K., Srinivasan, A., Banerjee, M., Ramasubramaniam, A.K., Köhler, J.R., Kadosh, D., Lopez-Ribot, J.L. Dispersion as an important step in the Candida albicans biofilm developmental cycle. PLoS Pathog. 6(3):e1000828 (2010).
-
Carlisle, P.L., Banerjee, M., Lazzell, A., Monteagudo, C., Lopez-Ribot, J.L. and Kadosh, D. Expression levels of a filament-specific transcriptional regulator are sufficient to determine Candida albicans morphology and virulence. Proc. Natl. Acad. Sci. USA 106: 599-604 (2009).
Cover article, see commentary: Bastidas, R.J. and Heitman, J., Proc. Natl. Acad. Sci. USA 106: 351-352 (2009). -
Banerjee, M., Thompson, D.S., Lazzell, A., Carlisle, P.L., Pierce, C., Monteagudo, C., Lopez-Ribot, J.L., and Kadosh, D. UME6, a novel filament-specific regulator of Candida albicans hyphal extension and virulence. Mol. Biol. Cell 19:1354-1365 (2008).
*both authors made equal contributions.
